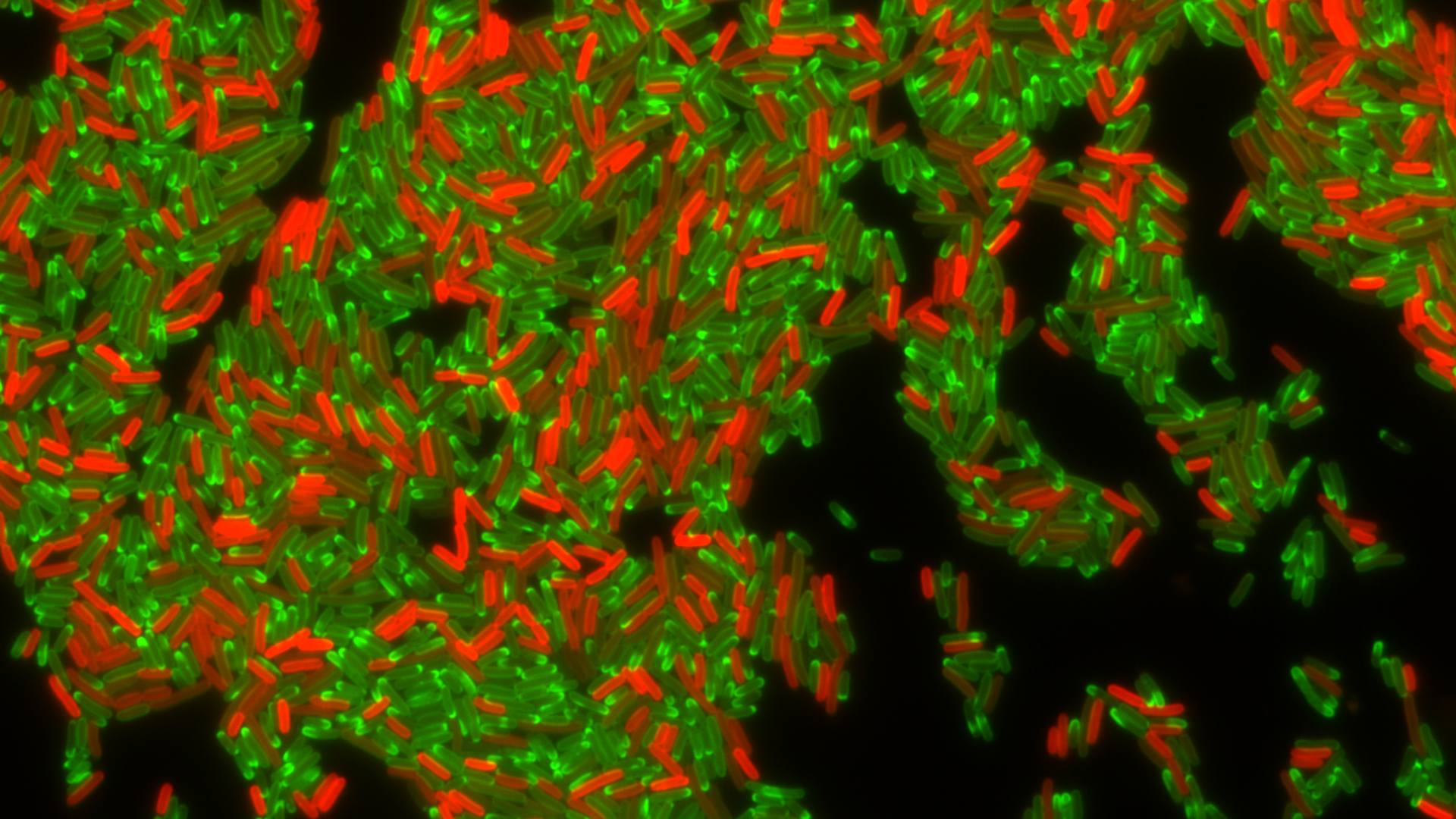
Drug-resistant bacteria producing a resistance factor, TetA efflux pump (in red), during treatment with tetracycline antibiotic (in green). Photo: Christian Lesterlin/MMSB department, University of Lyon, France
Using technology with real-time viewing, a team of scientists say in Science Thursday they can now show how quickly E. coli becomes resistant to tetracycline — finding that bacteria can pass genes with resistance to each other and then use a pump to keep most of the antibiotic out for the 2 hours it takes to render the previously sensitive bacteria resistant to the drug.
Why it matters: Antibiotic resistance is a growing threat that's projected to kill 10 million people every year by 2050. The discovery of how it occurs, at least in a lab setting, could help scientists develop an inhibitor that could be combined with an antibiotic to boost its effectiveness, study co-author Christian Lesterlin tells Axios.
What they did: During the 4-year project, the team first developed a system to monitor the transfer of DNA between cells.
- They added a resistance gene to the transferred DNA to look at how quickly resistance was established.
- Using live-cell microscopy, they tracked what the cells did in real-time. This was key to seeing how the process unfolded, says Lesterlin, who's a researcher at the University of Lyon in France.
- "With our system, we can look at things happening in real-time, describe the chronology, the time scale and learn about the genetic factors ... and the influence of antibiotics on this process," he says.
What they found, per Lesterlin:
- The bacterium can become resistant in less than 2 hours after getting the new genetic material: "It all happens very quickly."
- The role of the multidrug efflux pump is "essential" to the acquisition of new resistance by gene transfer between the E. coli.
- Even when the antibiotic tetracycline is present, the cells use the pump to export the antibiotics from the cell and "buy time for the cell" to acquire resistance.
- "The most surprising result was to observe that even in the presence of protein synthesis inhibitors (tetracycline antibiotic ), sensitive cells are still able to acquire the DNA, but more surprisingly they are still able to develop the resistance."
What they're saying: ETH Zurich's Vanessa R. Povolo and Martin Ackermann write in an accompanying perspective piece that the findings offer what could be an "important element" in the fight against antibacterial resistance — that the development of pump inhibitors will be key.
"Inhibitors of bacterial efflux pumps are receiving a lot of attention because they often make bacteria more sensitive to antibiotics. This study shows that these inhibitors could have a second, indirect, effect: They could block the transfer of resistance between bacteria during treatment. As a consequence, they could limit the emergence of new resistant strains," they write.
What's next: Lesterlin says their study is limited because it's only been conducted in the lab, so they hope to perform follow up tests in mammals, as well as testing other antibiotics or with other bacteria to see if they have similar responses. "Transferring this finding to the gut microbiota would be an important step forward."
Go deeper:
